DNA replication timing in humans
Accurate and efficient replication of the genome is a crucial prerequisite to successful cell proliferation, and defects in replication are associated with cancer, premature aging syndromes, and cell death. In eukaryotes, replication initiates at many replication origins across the genome, which fire at different times during the S phase of the cell cycle. This spatiotemporal organization of origin firing is termed the "DNA replication timing program" and has been associated with GC content, gene expression, chromatin structure, and local mutation rate.In my Ph.D. work, I examined several aspects of the replication timing program, each time with an eye toward method development:
Single-cell variation in replication timing
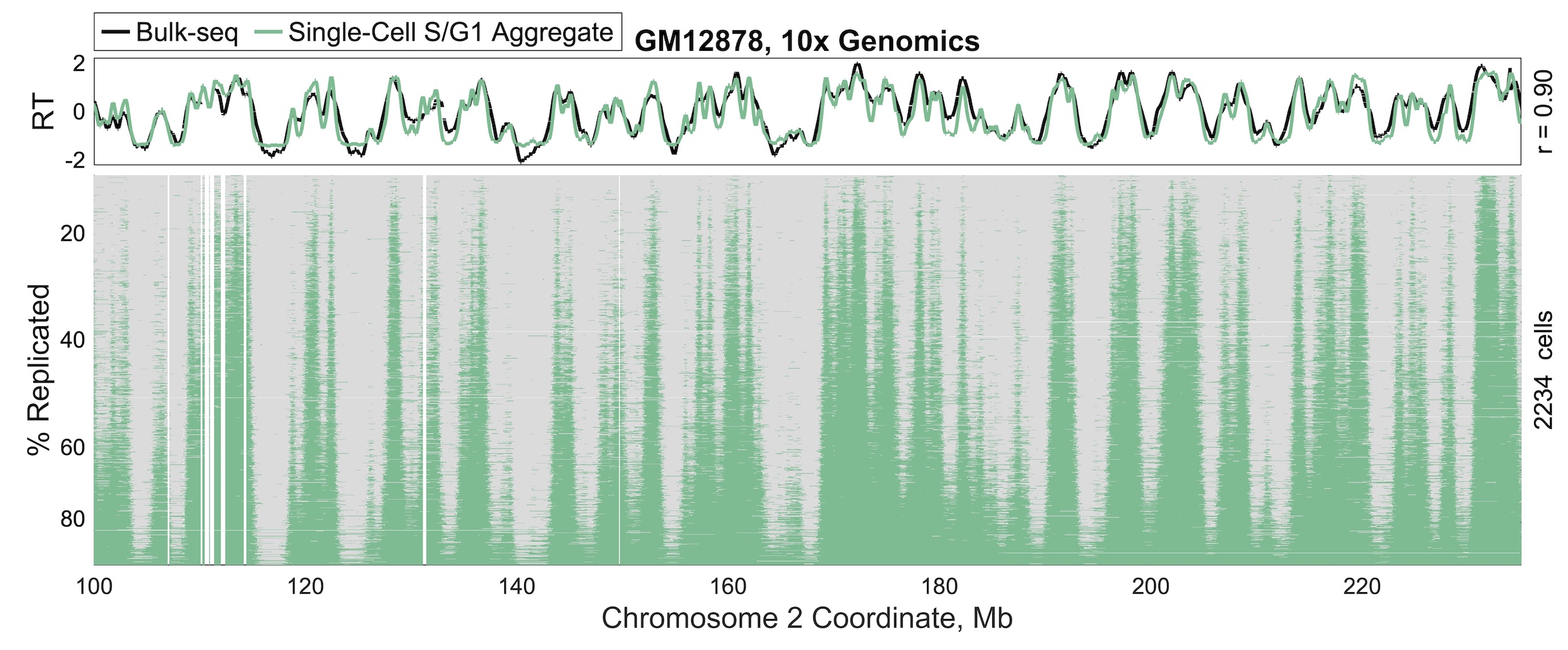
Replication timing has been observed to be highly reproducible across assays, yet the underlying mechanism(s) dictating its emergence are incompletely understood. To better parse out what kinds of mechanisms might be responsible, we developed an approach to assay replication timing in thousands of single cells. We find that replication timing is remarkably consistent across cells, but nonetheless, cell-to-cell variability is evident. This variability is most compatible with the notion that replication timing arises from disparate origin firing probabilities rather than from some kind of global "metronome". Read the paper in Nature Communications.
Inferring replication timing without cell sorting

Replication timing can be inferred from local fluctuations in read depth in whole-genome sequencing. However, a number of other factors (GC content, mappability biases, copy-number variations) can also impact read depth. Traditionally, these factors have been accounted for by sequencing a control sample of non-replicating (G1) cells in addition to a replicating (S-phase) sample. I contributed to the development of TIGER (Timing Inferred from Genome Replication), a pipeline for simulating a control sample to correct these confounding factors in a single unsorted sample. Read the paper in Bioinformatics.
Replication timing of human centromeres
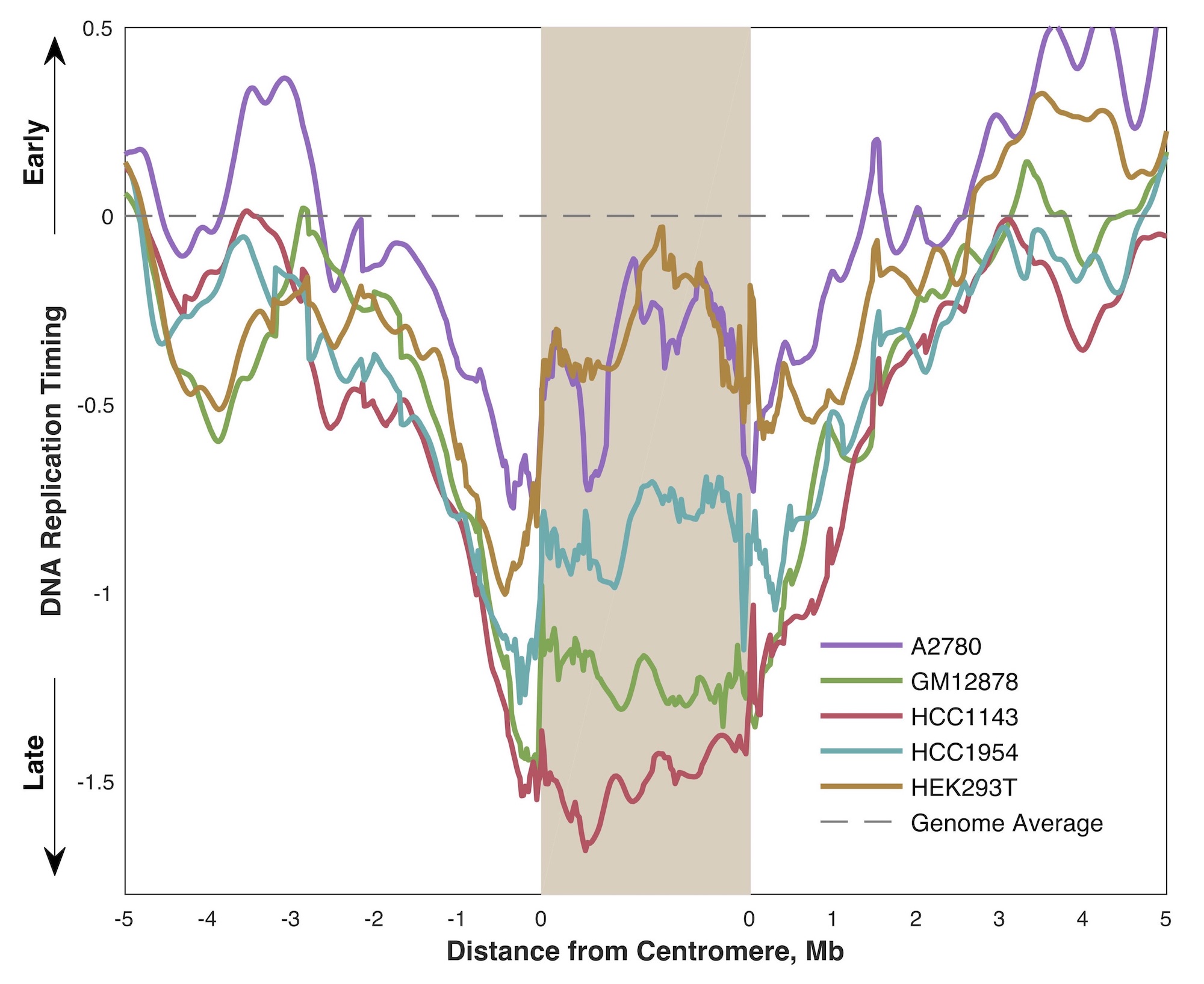
Centromeres are large, repetitive stretches of DNA that serve a critical structural role during cell division: the centromere is the attachment point for the mitotic spindle, which physically pulls the replicated sister chromatids apart into two daughter cells. Because of this repetitive sequence content, it is difficult to study centromere replication timing by sequencing. However, in collaboration with the Smolka lab, we showed that the centromeric model sequences included in the hg38 reference genome, coupled with paired-end sequencing, were sufficient to generate provisional replication-timing profiles for the majority of human chromosomes. In contrast to other heterochromatin (which replicates late in S phase), we find the centromeres replicate in mid-S phase and that centromeric replication timing is hyper-variable between cell lines, relative to other genomic regions. Read the paper in Genes.